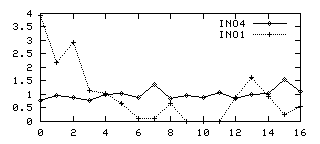
The Promoter of the Yeast INO4Regulatory Gene: a Model of the Simplest Yeast Promoter | Journal of Bacteriology
![PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a32978a889f29839465fc1784011fe6cbf49d278/9-Figure4-1.png)
PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar

IN02, A Positive Regulator of Lipid Biosynthesis, Is Essential for the Formation of Inducible Membranes in Yeast | Molecular Biology of the Cell

Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor | SpringerLink

The normalized expression values of INO2 and INO4 (INO-level) for each... | Download Scientific Diagram

Procedures of integrated analysis to reveal the effects of INO-level... | Download Scientific Diagram

Yeast transformation stress, together with loss of Pah1, phosphatidic acid phosphatase, leads to Ty1 retrotransposon insertion into the INO4 gene - Kudo - 2020 - The FASEB Journal - Wiley Online Library
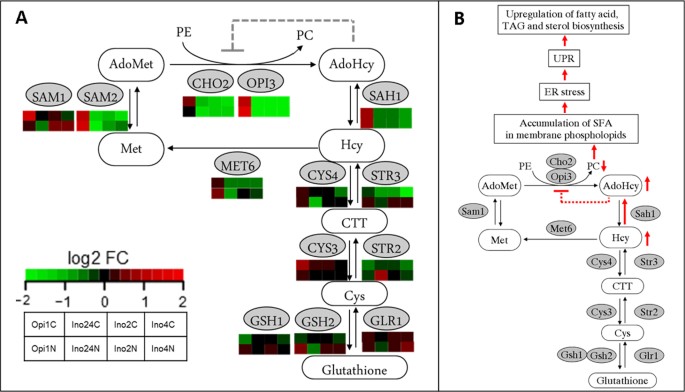
Integrated analysis, transcriptome-lipidome, reveals the effects of INO- level (INO2 and INO4) on lipid metabolism in yeast | BMC Systems Biology | Full Text

Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. - Abstract - Europe PMC
![PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a32978a889f29839465fc1784011fe6cbf49d278/10-Figure5-1.png)
PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar
Regulation of Inositol Metabolism Is Fine-tuned by Inositol Pyrophosphates in Saccharomyces cerevisiae*♦
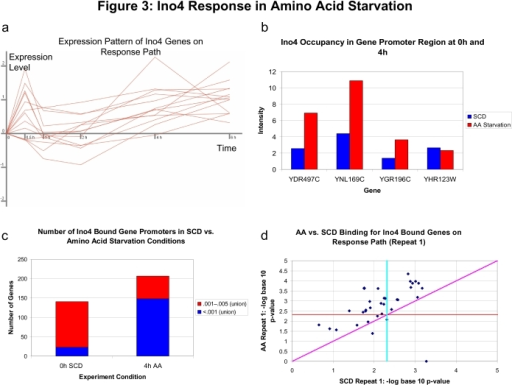
![PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a32978a889f29839465fc1784011fe6cbf49d278/8-Figure3-1.png)
PDF] High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae | Semantic Scholar

Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor | SpringerLink

Effects of the ino2, ino4, and opi1 mutations on the regulation of P... | Download Scientific Diagram

Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor | SpringerLink

The Promoter of the Yeast INO4Regulatory Gene: a Model of the Simplest Yeast Promoter | Journal of Bacteriology

Phosphatidic acid species 34:1 mediates expression of the myo-inositol 3-phosphate synthase gene INO1 for lipid synthesis in yeast - ScienceDirect

The normalized expression values of INO2 and INO4 (INO-level) for each... | Download Scientific Diagram